Zmora Lab -
Microbiome-Immune Interactions in Diseases of the Digestive System
R&D > Laboratories > Zmora Lab – Microbiome-Immune Interactions in Diseases of the Digestive System>>

Our Vision
The gastrointestinal tract harbors one of the highest densities of microorganisms on earth, with the number of bacteria outnumbering the host cells by a factor of 10 to 100. This complex microbial ecosystem, termed the microbiota, is separated from the host only by a layer of lining epithelial cells. The immune system is strategically located at the host-microbiota interface, where it can both regulate microbial ecology and sense microorganisms and their metabolites and translate those signals into physiological responses. This crosstalk between the immune system and the microbiota extends beyond the ability to maintain the balance between tolerance to commensal microorganism and immunity to pathogens, as it has been shown to affect various aspect of host physiology, including metabolism, autoimmunity and auto-inflammation, carcinogenesis and allergic reactions. In our lab we strive to unravel aspects of the host-microbiota crosstalk in gastrointestinal diseases and harness this knowledge to devise microbiota-based therapeutics. We use in-vivo and in-vitro models, complemented by microbiology, next-generation genomics and molecular biology techniques.

Contact Us
Primary Investigators

Dr. Niv Zmora , Lab PI
Phone: +972-3-6974281
Email: nivz@tlvmc.gov.il

Dr. Chen Varol , Co-PI
Phone: +972-3-6974226
Email: chenv@tlvmc.gov.il
Address
Research Center for Digestive Tract and Liver Diseases
Sourasky building
1st floor (above gate 6)
Sourasky Medical Center
6 weizmann st., Tel Aviv

Research
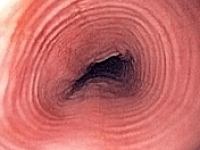
The rise in allergic diseases has been attributed to the ‘hygiene hypothesis’, according to which changes in the commensal microbiome in populations residing in industrial countries may trigger immune reactions. Indeed, associations have emerged between allergic diseases to variables that affect the microbiome assembly, such as history of cesarean delivery, premature birth,early antibiotic exposure and formula feeding. Eosinophilic esophagitis (EoE) is a chronic allergic disease, which causes significant impairment in the quality of life due to pain on swallowing, food impaction, esophageal stricture formation, and in extreme cases, esophageal rupture. Although previous literature has elucidated that EoE involves a breech to the gut barrier and activation of bacteria-sensing immune receptors, such as Toll-like receptors, only few studies have hitherto focused on the role of the microbiome in the disease. The association between EoE and an altered composition of the microbiome along the GI tract, from salivary through esophageal to fecal microbiome, has been shown repeatedly, however no insights into the cellular immune mechanisms that link between the microbiome to EoE development have been studied to date, partially owing to a lack of a reliable animal model for the disease.
In our lab we study the role of the microbiota in EoE using a unique mouse model for the disease developed by the Munitz lab (TAU) in various conditions, including treatment with antibiotics and under germ-free conditions, and attempt to ameliorate the disease using probiotic and post-biotic interventions. We employ an artificial esophagus culture system to systematically assess barrier function and the effect of microbiota- and host-derived compounds. We set out to validate our findings in human specimens by establishing an EoE human biobank, consisting of esophageal tissue biopsies, saliva and stool samples for microbiome analysis.
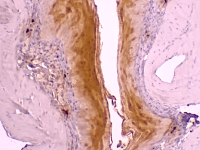
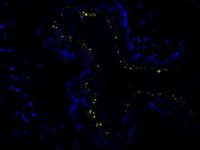
The alarmin S100A8/A9 is a heterodimer, also known as calprotectin, which is released from activated immune cells and keratinocytes and takes part in signal transduction pathways, which ultimately drive or suppress inflammatory responses. It has been implicated in many allergic diseases and shown to shape the intestinal microbiota in humans and mice, however despite mounting evidence, it has not been investigated to date in eosinophilic esophagitis (EoE). We set out to characterize S100A8/A9 in the esophagus at steady state and in the case of EoE and unravel clinical and immunological alterations in its absence. The overall goal of the study would be to utilize basic science and Next-Generation sequencing techniques to elucidate the role of S100A8/A9 in EoE and harness this knowledge to devise means to mitigate the disease. |
Eosinophilic esophagitis (EoE) is a rare, yet emerging, chronic type 2 immune-mediated disease, characterized by esophageal dysfunctions. It is likely to be primarily activated by food and air-born antigens. ADAMs (a disintegrin and metalloproteinases), are a family of more than 20 extracellular matrix (ECM) proteases that are released at sites of inflammation, where they selectively process ECM and cell surface molecules that further modulate immune cell migration and function. Among these, ADAM8 plays pivotal role in the development and propagation of type 2 allergic airway inflammation, but its role in EoE remains unknown. We intend to methodologically investigate the role of ADAM8 in different immune cells and epithelial cells in the development of experimental EoE. |
Gallery





Our Team
Current Staff
Research associate
- Noam Erez
Student
- Naama Sonigo
Current funding


Highlighted Publications
|
|
Zmora N, Suez J, Elinav E. Nat Rev Gastroenterol Hepatol. 2019 Jan;16(1):35-56. |
More Publications >>
Zmora N, Soffer E, Elinav E. Sci Transl Med. 2019 Jan 30;11(477).