Laboratory of Molecular Dermatology
R&D > Laboratories > Laboratory of Molecular Dermatology

Our Vision
Our laboratory has been investigating for the past 20 years the genetic basis of skin disorders. These efforts have led to the deciphering of the molecular basis of more than 25 genetic diseases by members of our group. The deciphering of the molecular basis of a monogenic disorder invariably reveals a novel pathway whose importance is exemplified by the disease resulting from its malfunction. Accordingly, over the years we have uncovered a number of novel biochemical pathways of importance for the physiology of the skin and other organs including kidney, eye and central nervous system. Once the function of a novel gene product is established, this new knowledge can be translated in the form of new treatments for rare and more common diseases alike.
Oscillating between bedside and the bench, our research program is substantiating a major goal of our division, the integration of research and clinical care to the benefit of our patients. Our research program's main objectives are:
• Delineation of the genetic basis of inherited skin disorders using a palette of analytical tools.
• Systematic investigation of the function of novel genes found to be associated with inherited skin diseases using humanized models such as three-dimensional artificial skin equivalents, hair organ cultures and chimeric mice.
• Translation of this new knowledge into treatments for rare and more common diseases using engineered reporter cell lines and robotic small molecule library screening followed by in vitro and in vivo validation of relevant hits.

Contact Us
Primary Investigators

Prof. Eli Sprecher, Lab PI
Department of Human Molecular Genetics & Biochemistry, Faculty of Medicine;Vice Dean for Clinical Affairs, Faculty of Medicine.03-6974287 elisp@tlvmc.gov.il

Dr. Ofer Sarig, Lab Manager
03-6973720 ofers@tlvmc.gov.il
Address
Founders Building
6th floor Room

Research
The epidermis, the outer layer of the skin, differentiates during a complex and tightly regulated process, involving the participation of no less than 400 distinct proteins. This process known as cornification culminates with the formation of the epidermal barrier, a complex of cells, proteins and lipids, which separates us from our immediate environment. Proper cornification is crucial for survival on the surface of the earth as attested by a high degree of conservation of this system among terrestrial animals as well as the catastrophic consequences of abnormal epidermal differentiation as exemplified by a large group of human diseases known as cornification disorders. The molecular diagnosis of these disorders is complicated by a high level of phenotypic and genetic heterogeneity and the patho-mechanisms underlying many of these disorders are largely unknown. In this project, we aim at expanding our understanding of the molecular mechanisms involved in normal epidermal barrier formation through the study of rare monogenic disorders of cornification and to translate the new knowledge in the form of new treatments for rare and more common diseases alike.
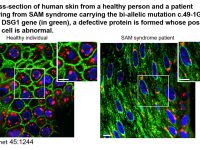
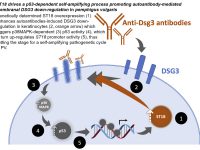
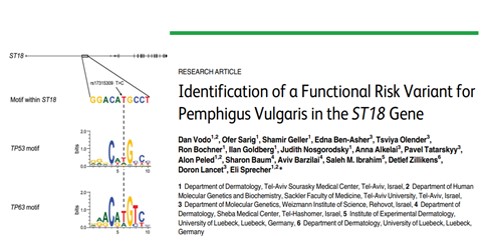
Pemphigus vulgaris (PV) is a life-threatening autoimmune skin blistering disease resulting from disruption of epidermal cell-cell adhesion. It is particularly prevalent among individuals of Jewish extraction. Treatment is focused on immunosuppression, exposing the patients to serious side-effects. We previously identified a functional PV-associated risk variant in the ST18 gene promoter, resulting in increased gene expression in patient skin, keratinocyte secretion of pro-inflammatory cytokines and destabilization of cell-cell adhesion, pointing to the possibility to intervene with the disease course by targeting cutaneous rather than immunological functions. In this project, we aim at further delineation the genetic basis of PV, investigate the pathways regulated by ST18 in PV and translate our findings into biomarkers and molecular targets for the treatment of PV.
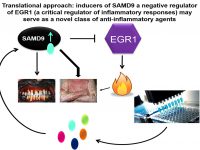
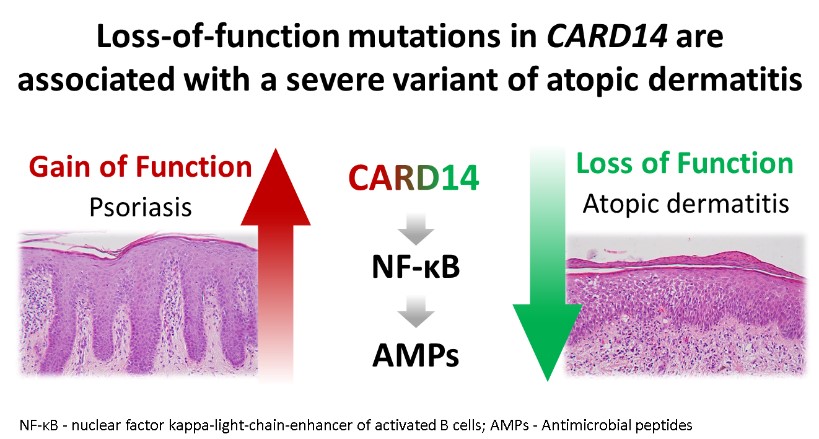
The deciphering of the molecular basis of a monogenic disorder invariably reveals a novel pathway whose importance is exemplified by the disease resulting from its malfunction. Accordingly, over the years we have uncovered a number of novel biochemical pathways of importance for the physiology of the skin and other organs. Once the function of a novel gene product is established, this new knowledge can be translated in the form of new treatments for rare and more common diseases alike. In this project we employing this strategy in the context of inherited inflammatory skin diseases based on our previous discoveries that defective function of SAMD9 or CARD14 are associated with aberrant activation of inflammation in the skin. We therefore reasoned that modulators of SAMD9 or CARD14 expression may serve as a novel class of therapeutic agents for skin inflammatory conditions. Thus, our goals in this project are to identify compounds capable of modulating SAMD9 or CARD14 expression using small molecules libraries screen and to dissecting out the mechanisms of action of those modulators.
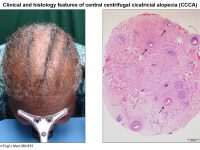
The deciphering of the genetic basis of monogenic disorders is not only beneficial in order to allow for proper genetic counseling and prenatal diagnosis to be provided to patients and their families, it also invariably uncovers unknown and very often unexpected aspects of our biology. Indeed, the consequences of the disease-associated protein’s malfunction, which are by definition evident in affected individuals, directly point to its physiological role. Over the past 20 years, our group has successfully been employing this strategy in the context of inherited skin diseases. The hair follicle is a complex mini organ of the skin. While hair research has developed dramatically in recent years, there are still very few treatments that can be offered to individuals suffering from hair disorders, e.g. hypotrichosis (baldness) or hypertrichosis and hirsutism (excessive growth of hair). Although compatible with normal lifespan, hair disorders affect many aspects of personal and social life. In this project we aim at investigate hair follicle biology through the study of rare inherited human hair diseases and translate the molecular knowledge gleaned to the development of novel therapies for those rare disease as well as for more common hear diseases (e.g. Central centrifugal cicatricial alopecia).
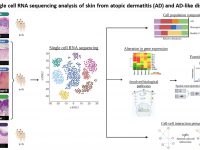
Atopic dermatitis (AD) is a chronic inflammatory disease affecting approximately 10-20% of the population worldwide. Interestingly, over the past few years, a number of monogenic disorders featuring AD-like clinical manifestations have been shown to result from defective function of epidermal adhesion proteins. These observations are in agreement with a growing body of evidence attributing to epidermal elements a primary role in the pathogenesis of AD, with obvious implications for the development of future therapies for this and other skin disorders. In this project, we aim at investigating the role of epidermal adhesion molecules in the context of activated inflammatory pathways in atopic dermatitis and other cutaneous disorders using a verity of standard as well as advance molecular techniques such as single cell RNA sequencing combined with spatial profiling for RNA and proteins.
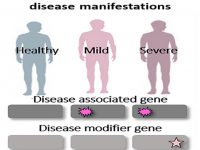
Even in the case of monogenic disorders which result from mutations within single and well-defined disease-causing genes, disease manifestations can vary dramatically. Whereas classical medical genetics has tended to distinguish between monogenic and multifactorial diseases, it is now increasingly evident that human diseases exist on a continuum between these two ends. Indeed, even siblings carrying the same mutation, occasionally display extremely divergent phenotypes, sometimes ranging between life-threatening phenotypes to absence of phenotype. This divergence is often hypothesized to result from genetics modifiers and/or epigenetic mechanisms such us DNA methylation and micro-RNAs post-transcriptionally regulation that control disease manifestations. To study this phenomenon in skin diseases, we are working on deep phenotypic and functional characterization of monogenic skin diseases to broaden our understanding of the pathogenesis and clinical manifestations of these diseases as well as to develop innovative approaches for diagnosis and treatment of those diseases.
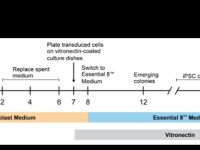
Drug discovery remains an unprecedented challenge because of cost, time, and high failure rates in clinical trials. Part of these difficulties stems from the fact that in many cases, animal models fail to accurately reflect human physiology and to recapitulate patient-specific features. In-vitro models in two or three dimensional cell cultures using standard molecular biology methodologies (e.g. silencing or over expressing gene of interest, induce or inhibit molecular pathways etc.) offer the advantage of simplicity and cost-effectiveness, but in many cases also fail to exhibit patient-specific features. Patient-derived induced pluripotent stem cells (PD-iPSCs) technology compensate for many of the above disadvantages. iPSCs generated from individual somatic cells offer an unlimited proliferation capacity and carry the exact genetic background including disease causing and unknown modifier genes and have the remarkable ability to recapitulate in vitro the main steps of skin development and homeostasis and provide an extremely useful option to generate patient-specific organotypic diseased skin. This makes PD-iPSCs an ideal tool to explore the molecular mechanisms underlying different skin disorders and consequently the development of drug screening platforms. Using this platform, we are currently developing novel therapeutic approach for the treatment of atopic dermatitis and Netherton syndrome.
Gallery










Our Team
Current Staff
Physicians-scientists
- Dr. Sari Assaf
- Dr. Avital Baniel
- Dr. Danny Daniely
- Dr. Janan Mohamad
- Dr. Mor Pavlovski
- Dr. Alon Peled
- Dr. Liat Samuelov
- Dr. Dan Vodo
- Dr. Livia Preisle
- Dr. Carmel Bilu
- Dr. Lior Lasman
- Dr. Noy Keller
MD students
- Erel Geller
PhD students
- Shir Bergson
MD PhD students
- Yarden Feller
- Kiril Malovitski
Technician
- Odile Meijers
- Noy Eretz Kdosha
Past Staff
Alumni
- Judeh Abu Sa’d
- Ron Bochner
- Ilana Chefetz
- Dan Ciubotaru
- Ori Eytan
- Gilad Fainberg
- Levi Fried
- Dana Fuchs-Telem
- Dalit Ganani
- Meital Grafi-Cohen
- Dov Hershkovitz
- Margarita Indelman
- Shirli Israeli
- Jennie Lugassy
- Lee Magal
- Natalia Malchin
- Liron Malki
- Mordechai Mizrachi-Koren
- Sagi Nahum
- Janna Nousbeck
- Gilly Padalon-Brauch
- Tova Rogers
- Shahar Taiber
- Orit Topaz
- Emily Warshauer
- Hadas Zamir
- Valeria Briskin
Current funding









Active Collaborations
- King’s college, United Kingdom
- Dundee university, Scotland
- Thomas Jefferson University, USA
- Northwestern University, USA
- Bonn University, Germany
- St Georges hospital, Australia
- Luebeck University, Germany
- Kirov State Medical Academy, Russia
- University of Manchester, United Kingdom
- Necker Children's hospital and Institut Imagine, France
- Stanford University, USA
- University of KwaZulu–Natal, South Africa

Highlight Publications
Cutaneous and Developmental Effects of CARD14 Overexpression in Zebrafish.
Baniel A, Ziv L, Ben-Moshe Z, Sarig O, Mohamad J, Peled A, Rechavi G, Gothilf Y, Sprecher E. Biomedicines. 2022
Autosomal recessive congenital ichthyosis caused by a pathogenic missense variant in CLDN1.
Mohamad J, Samuelov L, Assaf S, Malki L, Malovitski K, Meijers O, Adir N, Granot E, Pavlovsky M, Sarig O, Sprecher E. Am J Med Genet A. 2022
A unique skin phenotype resulting from a large heterozygous deletion spanning six keratin genes.
Mohamad J, Sarig O, Beattie P, Malovitski K, Assaf S, O’Toole E, Schwartz J, Evans H, Samuelov L, Sprecher E. Br J Dermatol. 2022
Assaf S, Vodo D, Malovitski K, Mohamad J, Bergson S, Feller Y, Malki L, Sarig O, Sprecher E. Sci Rep. 2022
Loss-of-function variants in KLF4 underlie autosomal dominant palmoplantar keratoderma.
Malovitski K, Sarig O, Assaf S, Mohamad J, Malki L, Bergson S, Peled A, Eskin-Schwartz M, Gat A, Pavlovsky M, Sprecher E. Genet Med. 2022
Mohamad J, Samuelov L, Malchin N, Rabinowitz T, Assaf S, Malki L, Malovitski K, Israeli S, Grafi-Cohen M, Bitterman-Deutsch O, Molho-Pessach V, Cohen-Barak E, Bach G, Garty BZ, Bergman R, Harel A, Nanda A, Lestringant GG, McGrath J, Shalev S, Shomron N, Mashiah J, Eskin-Schwartz M, Sprecher E, Sarig O. Exp Dermatol. 2021
Loss-of-function variants in C3ORF52 result in localized autosomal recessive hypotrichosis.
Malki L, Sarig O, Cesarato N, Mohamad J, Canter T, Assaf S, Pavlovsky M, Vodo D, Anis Y, Bihari O, Malovitski K, Gat A, Thiele H, White BEP, Samuelov L, Nanda A, Paller AS, Betz RC, Sprecher E. Genet Med. 2020
Loss-of-Function Variants in SERPINA12 Underlie Autosomal Recessive Palmoplantar Keratoderma.
Mohamad J, Sarig O, Malki L, Rabinowitz T, Assaf S, Malovitski K, Shkury E, Mayer T, Vodo D, Peled A, Daniely D, Pavlovsky M, Shomron N, Samuelov L, Sprecher E. J Invest Dermatol. 2020
More Publications >>
Variant PADI3 in Central Centrifugal Cicatricial Alopecia.
Malki L, Sarig O, Romano MT, Méchin MC, Peled A, Pavlovsky M, Warshauer E, Samuelov L, Uwakwe L, Briskin V, Mohamad J, Gat A, Isakov O, Rabinowitz T, Shomron N, Adir N, Simon M, McMichael A, Dlova NC, Betz RC, Sprecher E. N Engl J Med. 2019
Peled A, Sarig O, Sun G, Samuelov L, Ma CA, Zhang Y, Dimaggio T, Nelson CG, Stone KD, Freeman AF, Malki L, Vidal LS, Chamarthy LM, Briskin V, Mohamad J, Pavlovsky M, Walter JE, Milner JD, Sprecher E. J Allergy Clin Immunol. 2019
Mohamad J, Sarig O, Godsel LM, Peled A, Malchin N, Bochner R, Vodo D, Rabinowitz T, Pavlovsky M, Taiber S, Fried M, Eskin-Schwartz M, Assi S, Shomron N, Uitto J, Koetsier JL, Bergman R, Green KJ, Sprecher E. J Invest Dermatol. 2018
Bochner R, Samuelov L, Sarig O, Li Q, Adase CA, Isakov O, Malchin N, Vodo D, Shayevitch R, Peled A, Yu BD, Fainberg G, Warshauer E, Adir N, Erez N, Gat A, Gottlieb Y, Rogers T, Pavlovsky M, Goldberg I, Shomron N, Sandilands A, Campbell LE, MacCallum S, McLean WHI, Ast G, Gallo RL, Uitto J, Sprecher E. J Invest Dermatol. 2017
Peled A, Sarig O, Samuelov L, Bertolini M, Ziv L, Weissglas-Volkov D, Eskin-Schwartz M, Adase CA, Malchin N, Bochner R, Fainberg G, Goldberg I, Sugawara K, Baniel A, Tsuruta D, Luxenburg C, Adir N, Duverger O, Morasso M, Shalev S, Gallo RL, Shomron N, Paus R, Sprecher E. PLoS Genet. 2016
Ustekinumab therapy for Netherton syndrome.
Samuelov L, Shehadeh W, Sarig O, Gat A, Matz H, Sprecher E.
J Dermatol. 2023
Identification of a Functional Risk Variant for Pemphigus Vulgaris in the ST18 Gene. Vodo D, Sarig O, Geller S, Ben-Asher E, Olender T, Bochner R, Goldberg I, Nosgorodsky J, Alkelai A, Tatarskyy P, Peled A, Baum S, Barzilai A, Ibrahim SM, Zillikens D, Lancet D, Sprecher E. PLoS Genet. 2016 May 5;12(5):e1006008. doi: 10.1371/journal.pgen.1006008. eCollection 2016 May. PMID: 27148741 Free PMC article. |